In this lab, we studied the uses of the compound light microscope by analyzing hair, string, magazine print, and a ruler with the microscope and with the naked eye. The purpose of this lab was to explore visualization of common items with the microscope. The microscope is capable of enlarging objects by factors of 10, allowing for details that are invisible to the naked eye to become apparent. While completing this lab, we learned about the parts of a microscope and their functions, the process of making a wet slide, and the change in orientation of an image when viewed in a microscope.
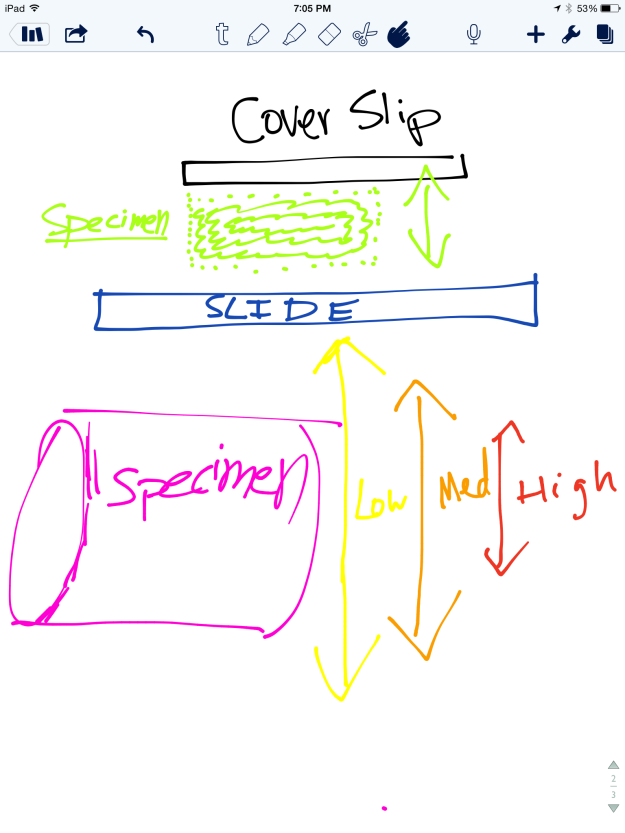
As the human eye cannot distinguish objects smaller than 0.1 millimeters, the microscope is a helpful tool for magnifying specimens. There are two common types of microscopes: light and electron. Light microscopes use light waves to illuminate the object being examined, while electron microscopes use a series of electrons to visualize the object. I used a light microscopes, which is often used for general laboratory work, to magnify hair, string, magazine print, and a ruler. Light microscopes can magnify objects from 40 to 1000 times, but we only measured data at 40X and 100X. The specimens were prepared on a slide using the wet mount method, which is the most frequently used form of slide preparation. Below is a diagram of how light travels in a light microscope from the specimen being examined to the eye.
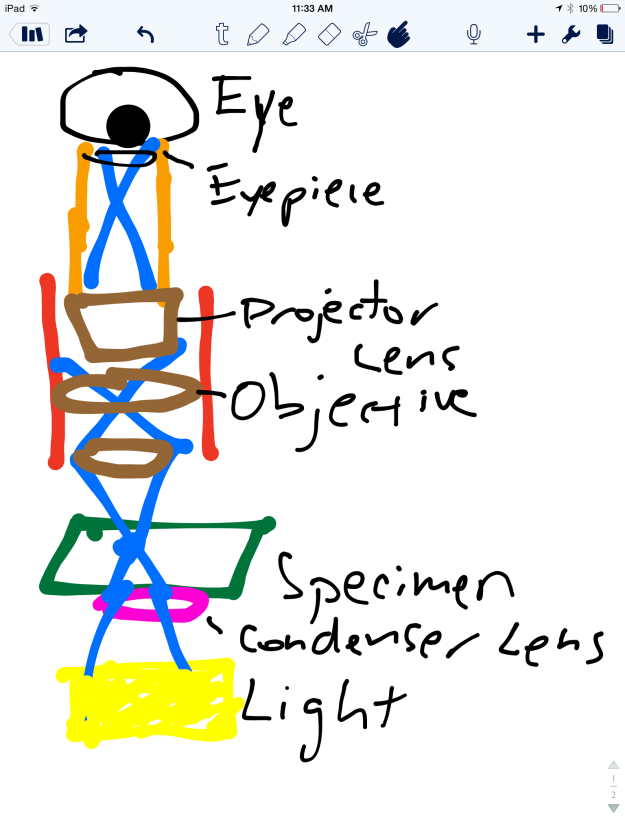
For this lab we examined the following items:
-magazine or newspaper clippings
-colored threads
-single strand of hair from two individuals with different hair color.
-transparent ruler
We used the following materials to prepare the slides and see the magnified images:
-light microscope with 10X eyepiece, and 4X and 10X objective lenses attached.
– three glass slides
– three coverslips
-water
-plastic pipette
-scissors
Setting up the microscope:
1. I clicked the the low power (4X) objective lens into position.
2. I opened the diaphragm fully.
3. I turned on the microscope light switch.
Preparing the newspaper slides:
1. I cut out a lowercase “o” from the clipping.
2. I placed the o slip facing down on the slide.
3. A drop of water was added to the slide, which turned the paper face up.
4. I gently placed the coverslip atop the o.
5. The process was repeated with lowercase “e” and “r”.
Preparing the hair and string slides:
1. The strands of hair were placed in a crossing position.
2. A drop of water was added to the slide.
3. The coverslip was placed above the cross.
Using the microscope on newspaper slides:
1. The “o” slide was placed on the stage.
2. The microscope was focused using the coarse and fine adjusting knobs.
3. After viewing on 4X, I looked at the o using the 10X lens.
4. I repeated the steps for the “e” and “r” slides.
Using the ruler:
1. The ruler was positioned on the stage so it covered half of the light opening.
2. The ruler was viewed using the 4X and 10X objective lenses.
Using the hair and string:
1. The slide with hair was placed on the stage, with the cross visible through the eyepiece.
2. I observed the slide using the 4X and 10X objective lenses.
3. This was repeated using the string slide.
I noted my observations after viewing all of the slides.
Note: If the mount became dry while viewing, I added a drop of water under the coverslip to improve visualization.
Fig 1:
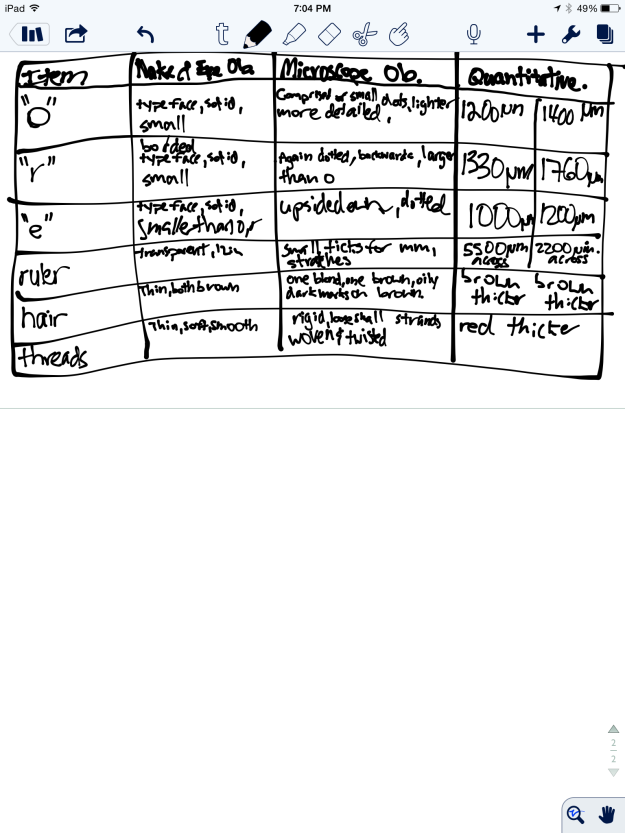
Figure 1 depicts my observations, including qualitative observations with the naked eye and the microscope, as well as estimated widths of the specimens.
Fig 2:
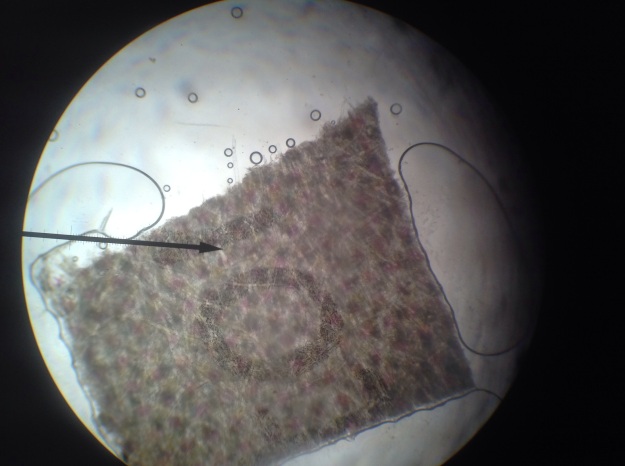
Figure 2 depicts the “o” magnified by a factor of forty. As noted in Figure 1, the o appears to be made of dots, and the estimated width is 1200 microns.
Fig 3:
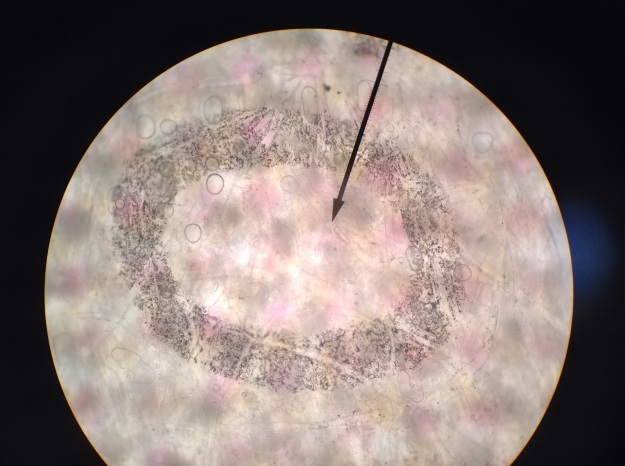
In Figure 3, the dots appear larger, as the specimen has been magnified by 100X. The estimated width in this image is 1400 microns.
Fig 4

Figure 4 illustrates the “e” at 100X magnification. It appears grainy in texture, and the estimated width is 1200 microns.
Fig 5
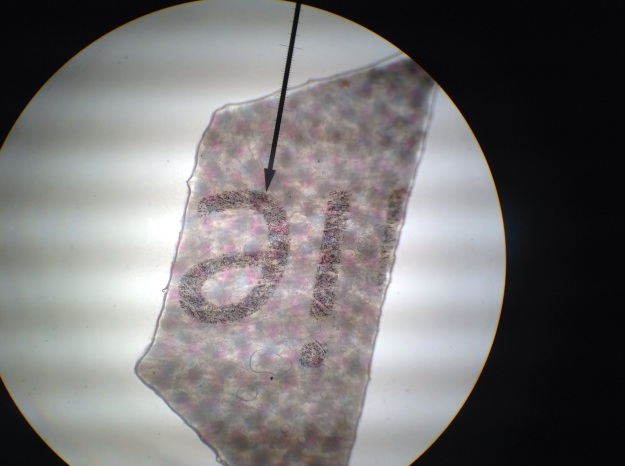
Figure 5 depicts the “e” at 40X magnification. The estimated width is 1000 microns, and the neighboring character is still visible.
Fig 6

Figure 6 shows the “r” at 100X magnification. It appears to be made of dots, and it appears larger than the e and o. The estimated width is 1760 microns.
Fig 7

Figure 7 illustrates the cross of the strings at 100X magnification. The strings are estimated to be 600 microns across.
Fig 8

Figure 8 depicts the three threads at 40X magnification. Both the twisted structure of the thread and additional loose strands are visible, and the estimated width is 400 microns.
Fig 9
Figure 9 depicts two hairs at 40X magnification. The lighter hair is blond, while the darker hair is brown. The brown strand is slightly thicker than the blond hair: the blond hair is about 54 microns across, and the brown hair is about 62 microns across.
Scratches were visible on the surface of the ruler in addition to the 5.5 millimeters in the field of view.
In this lab, we explored the function and uses of a compound light microscope. The microscope has a light below the stage that passes through the specimen and then is refracted by two lenses in a manner that magnifies the specimen.
We observed 6 specimens: hair, string, letters r, e, and o, and the ruler.
*As described in the table, there were several differences and similarities between an image viewed through a microscope and the same image viewed with the naked eye.
With the naked eye, the typeface letters e, r, and o appear to be correctly oriented and facing up. In the microscope, the e and r appeared backwards and upside down. This occurs because the objective and ocular lens are convex. The light rays converge together at a central point: the focal point. When the image is magnified, the objective lens has a short focal length and an upside down image is made (see figure 10 below). This image is then magnified further through the eye piece. The letter e was useful demonstrating this effect, as with the letter o it was harder to see that the image was inverted and upside down.
Figure 10
Images appear smaller and inverted when light is focused beyond the lens focal point.
When observed through the microscope, the hair, ruler, and string had minute details that were not apparent with the naked eye. For example, the ruler had fine scratches, and small details like oil and color were apparent when examining the hair. I also noted both the twisted structure of the string and small threads coming out of its main strand.
*When viewing an object through the higher powered objective, not all of the object was in focus. This was most apparent when I observed the overlapping hair and thread. There are 3 reasons for this observation. First, the field of view is largest at low power. This is the best power to locate the specimen. Second, the depth of focus decreases at higher magnifications. Third, at higher power the amount of light is reduced. This diminishes resolving power or the ability to see two close objects as separate items. As we observed the different colored threads, the top thread was in focus, then the middle thread, then the lowest thread. They were not sharply in focus at the same time. The fine-adjustment knob helped focus on different layers of depth, but as one thread came into focus the others went out of focus. This is illustrated below.
Fig 11
At higher magnifications the depth of focus decreases
*The order of the overlapping colored threads was black over red over green.
*There is an inverse relationship between the total magnification and the diameter of the field of view. As the magnification increases, the diameter proportionally decreases. This can be further illustrated by the following equation:
diameter of field B = magnification of A X diameter of A
————————————————
magnification of B.
Where a is any magnification at which the diameter of the field is known, and B is any magnification at which the diameter of the field is unknown.
I also observed that moving the slide to the right, made the image move to the left and moving the slide away from me, made the image move towards me.
*The diameter in micrometers of the low power field of view of my microscope was approximately 5.5 mm, or 5500 micrometers.
*To calculate the diameter in micrometers of the high power field, I can use the following equation:
magnification number of higher-power objective
————————————————————— = A
magnification number of lower-power objective
diameter of low-power field of view
—————————-–——————–
A. =diameter of high -powered field of view
The value of A is 10/4, or 2.5. The diameter was found to be 5500 micrometers, when divided by 2.5 equals 2200 microns, the diameter of the high powered field of view.
*The diameter of the high power field was 2200 microns. The hair is about one 31st of the screen, which corresponds to an estimated width of 70 microns.
One problem that I had during the experiment was adjusting the diaphragm to provide even illumination in the photographs. If I were to repeat this experiment, I would like to explore in more depth the inversion of the magnified objects. I would replace the eye piece with another convex lens to see if I could maintain the magnification, but correct the orientation of the observed specimen to match the true specimen. I would also attempt to improve the depth of focus at higher magnification. Next, I would like to determine if varying the light intensity would help with visualization. I would then examine human cells under the microscope, possibly by preparing a wet mount of cells scraped gently from the inside of my cheek. Finally, I would compare a specimen under a light microscope with the images obtained using an electron microscope and a fluorescence microscope.
References:
cas.miamioh.edu/mbiws/microscopes/compoundmicroscope.html
abacus.bates.edu/~ganderso/biology/resources/microscopy.html






